Results on its interaction with antibiotics may lead to the development of new forms of treatment for this disease
Tuberculosis is a chronic infection usually caused by a bacterium called Mycobacterium tuberculosis. This bacterium infects cells of the immune system called alveolar macrophages, which are responsible for removing pollutants and microorganisms from the surface of the alveoli, where the exchange of gases occurs during respiration.
It is estimated that approximately two billion people worldwide are infected with M. tuberculosis without symptoms. However, the clinical manifestations of the disease may appear at any time in life, especially when the immune system is weakened, such as due to malnutrition or diseases such as cancer and AIDS.
Tuberculosis is considered a curable disease when the patient is diagnosed and treated promptly with antibiotics. Nevertheless, the chronicity of this infection makes it difficult to eradicate bacteria altogether. Generally, patients must take the medication for several months, making it harder for them to persist in the treatment and favoring the emergence of antibiotic-resistant bacteria. In recent years, the emergence of new bacteria, resistant to routine treatments, has been a worldwide concern and it is imperative to seek new therapeutic strategies against this disease.
The cell wall is an essential structure for the life of most bacteria. It surrounds the cell membrane of these microorganisms and is formed by a complex entanglement of lipids and carbohydrates that mainly provide protection to the bacterial cell. A set of enzymes called L,D-transpeptidases are involved in the formation of this shield. Therefore, inhibition of these enzymes could lead to the death of bacteria or at least facilitate the action of our body’s defense cells.
An important group of antibiotics are the so-called beta-lactam antibiotics. This group includes antibiotics such as penicillin and its derivatives. Another class included in this group are carbapenems, antibiotics capable of resisting bacterial enzymes that inhibit the action of penicillin.
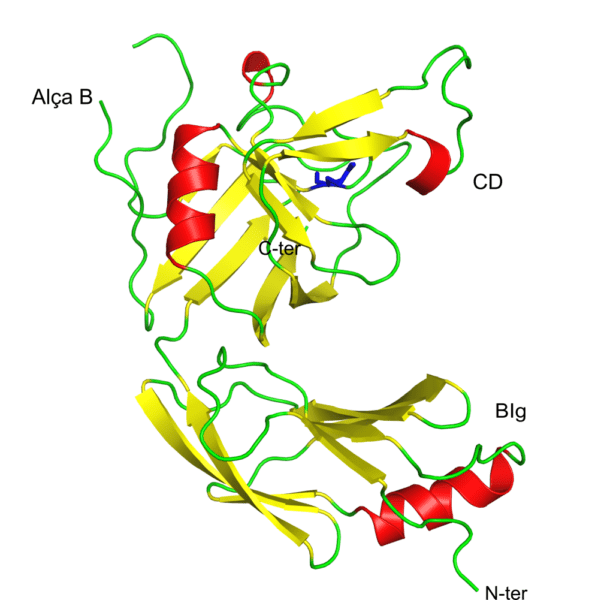
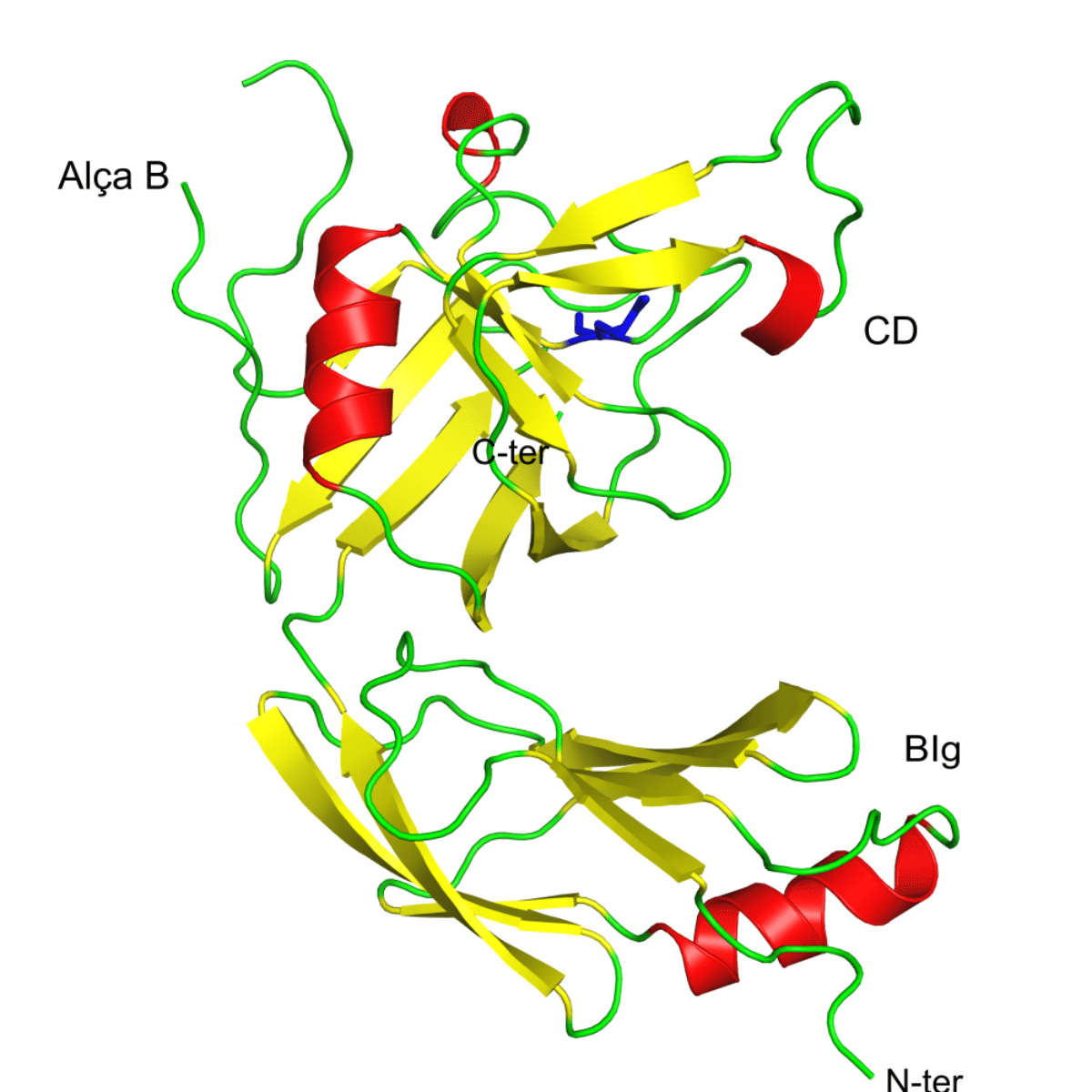
Elements of the secondary structure of L,D-transpeptidase-3 from Mycobacterium tuberculosis acylated by an acetyl fragment derived from faropenem. Beta sheets in red, α-helices in yellow and the loops are shown in green. The figure shows, at the amino terminus (N-ter), the bacterial domain similar to immunoglobulin (BIg) and in the carboxy terminus the catalytic domain (CD). B-loop is a unique structure of this enzyme when compared to the other M. tuberculosis L,D-transpeptidases. In blue is shown an acetyl fragment covalently attached to cysteine 246 at the active site of the enzyme. Figure taken with Pymol.
Some studies have shown that carbapenems are capable of inhibiting L, D-transpeptidases, even though they are not used in tuberculosis therapy. Thus, knowledge of the mechanisms of interaction between L,D-transpeptidases and these drugs may lead to the improvement of existing beta-lactams or to the development of new beta-lactams that may be useful for the treatment of tuberculosis variants which are resistant to other antibiotics.
In a recent publication in the journal ACS Infectious Diseases [1], Gerardo A. Libreros-Zúñiga et al. determined the structure of L,D-transpeptidase-3, one of the five enzymes of this family present in Mycobacterium tuberculosis.
The structure of this enzyme was investigated both in its isolated protein form (apo) and inactivated by beta-lactam faropenem, in addition to identifying affinities for nine other beta-lactams and to propose how these antibiotics could bind to the enzyme. The research thus provides information that can be used in the development of new drugs that act on L,D-transpeptidases and that may contribute to the treatment of tuberculosis.
According to the researchers, among the different experimental approaches used, the MX2 macromolecular crystallography beamline of the Brazilian Synchrotron Light Laboratory (LNLS) was a key tool to determine the enzymatic structures of the L,D-transpeptidase-3 enzyme in high resolution, in addition to making it possible to observe the degradation of the faropenem molecule by the enzyme.
The research was carried out with support from FAPESP – Sao Paulo Research Foundation (Grants 2015/0918808 and 2010 / 15971-3).
Source: [1] Structural Basis for the Interaction and Processing of β-Lactam Antibiotics by l,d-Transpeptidase 3 (LdtMt3) from Mycobacterium tuberculosis. Gerardo Andrés Libreros-Zúñiga, Catharina dos Santos Silva, Rafaela Salgado Ferreira, and Marcio Vinicius Bertacine Dias, ACS Infectious Diseases. DOI: 10.1021/acsinfecdis.8b00244
Study contributes to the understanding of mechanisms involved in neurodevelopmental disorders
Research investigates material based on poly(butylene succinate), urea and clay