These observations open up possibilities for the study of protein folding and provide a new interpretation to explain the nature of the cooperative behavior of proteins during folding reactions.
It is known that proteins are far from equilibrium during folding reactions, and they undergo a wide range of conformational states to reach the global folding minimum. Various physical and chemical strategies, such as the use of high temperature, high pressure, protonation, altered ionic strength, and harsh de- naturants, are commonly used to disturb folding species to promote the formation of rarely observed folding intermediates.
From the thermodynamic point of view, any perturbing agent affecting protein folding is controlled by Le Chatelier’s principle. For instance, increasing the concentration of urea shifts the folding equilibrium toward the unfolded state because of the increased preferential binding of urea to this state. In the case of pressure, the smaller volume of the unfolded state is favored at high pressure because it only affects the volumetric properties of the molecule.
The energy landscape theory of folding assumes that protein folding is the progressive organization of an ensemble of partially folded structures through which the protein passes on its way to a native conformation. Accordingly, proteins have a rugged funnel-like landscape that is biased toward their native structure due to evolution. Obtaining structural and dynamic information on multiple-stage protein intermediates that follow the folding trail may increase our knowledge of protein misfolding, which is associated with amyloidosis, prion formation, and the occurrence of several diseases, including Parkinson’s disease, Huntington’s disease, spongiform encephalopathy, and, more recently, cancer.
When using physical or chemical perturbations, NMR spectroscopy is a powerful tool to reveal a shift in the native conformation toward local intermediates that act as seeds for misfolding. The application of pressure to a protein forces the water shell into the protein, thus shifting the equilibrium of the system from the native state to an intermediate or to the unfolded state. Pressure appears to favor water infiltration into the protein and the disassembly of protein cavities, leading to increased hydration and decreased partial molar volume.High pressure can be coupled with various by various techniques but to better understand local changes, high-pressure NMR (HP-NMR) is adopted as the most informative approach. SAXS is another useful tool for assessing changes in protein folding and the size and shape of macromolecules in solution.
The binding of urea to proteins is based on the ability of urea to displace water molecules from the first solvation shell, thus increasing the system entropy and weakening the hydrophobic effect. Urea is also believed to form hydrogen bonds with the amide unit of peptide bonds.
Two main theories exist to explain why urea induces protein denaturation: The first hypothesizes an indirect mechanism by which urea alters the water structure and leads to hydrophobic group solvation; The second view is based on the direct interaction of urea with the protein. Most studies are based on model compounds and theoretical and modeling approaches, such as molecular dynamics.
Recently, Guilherme A. P. de Oliveira and Jerson L. Silva have studied Moniliophthora perniciosa necrosis- and ethylene-inducing protein 2 (MpNep2), shown in Fig 1. They systematically studied protein folding in response to chemical (urea), physical (pressure), or both perturbations using HP-NMR spectroscopy and 3D low-resolution shape reconstructions from SAXS data to better understand how each of the two disturbing agents (pressure and urea) affect protein folding and to examine how urea denatures proteins. The use of these synergistic techniques (HP-NMR and SAXS) provides reliable structural information on local/global intermediates within protein energy landscapes. They provide evidence of a conformational state in which urea binds to the protein without promoting full denaturation by populating a dry molten globule (DMG).
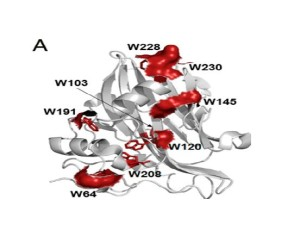
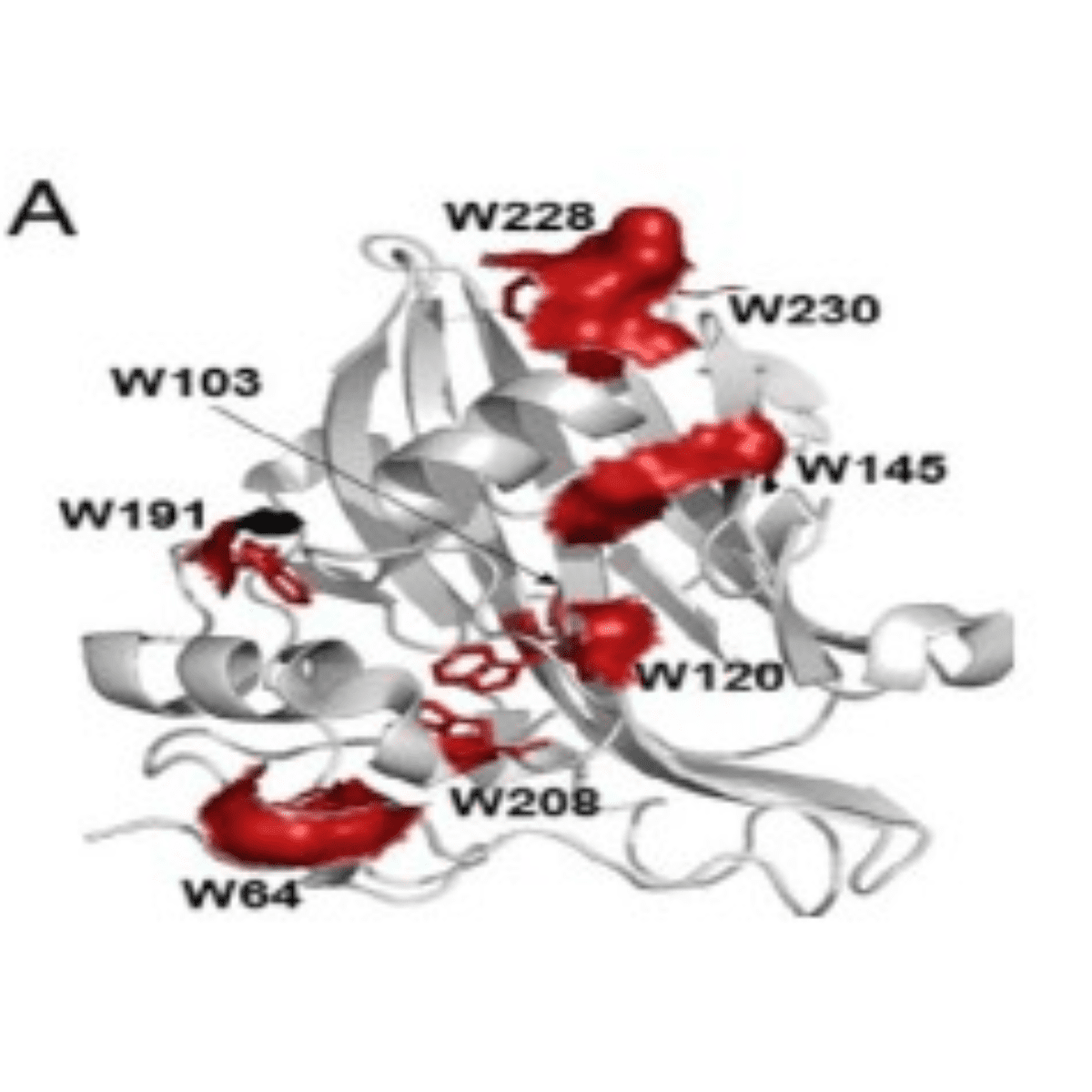
Figure 1. (A) A schematic representation of the secondary structural elements of MpNep2 (gray). Trp side chains are highlighted as red sticks, and the corresponding solvent-accessible surfaces of these residues are shown.
Based on their NMR, SAXS (SAXS data were collected using the SAXS1 small-angle scattering beamline of the National Synchrotron Light Laboratory, Campinas, Brazil), and Steady-state fluorescence spectroscopy results, the authors present the “push-and-pull” hypothesis to account for the effects of chemical and physical (pressure) denaturation of the folded state of a protein. Although both chemical and pressure denaturation processes are controlled by Le Chatelier’s principle, the molecular mechanisms are different. It is known that pressure shifts the equilibrium toward the denatured state due to the negative volume change, which is generally explained by a combination of several effects: the disruption of water-excluded cavities in the protein interior, electrostriction of broken salt bridges, and general water solvation. Monitoring each amino acid residue by NMR has permitted to map the progressive effects of pressure. At low pressures, linear effects are observed as pressure is increased due to the shortening of hydrogen bonds. The population of these low-lying excited states is a springboard to partially unfolded states. Thus, pressure leads to a homogeneous shrinking of the protein, followed by an unfolding intermediate with part of the hydrophobic core exposed and, ultimately, full denaturation (Fig 2).
In contrast, urea denaturation does not affect protein conformation until cooperative unfolding to the denatured state occurs. The “push-and-pull” mechanism has a dual character, explaining the general differences expected after denaturation with pressure vs. chemical agents. The use of a two-state assumption is the simplest thermodynamic analysis for both urea and pressure effects, and the analysis would be little affected if more complex assumptions were used.
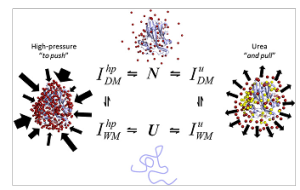
Fig. 2. Urea and high pressure results in different local unfolding states. The ensemble of dry (DM) and wet molten globule (WM) intermediates (I) in response to high pressure and urea are different according to NMR spectroscopy. A schematic of the “push-and-pull” hypothesis used to explain the mechanisms of the physical and chemical unfolding of proteins. Red spheres indicate water molecules, and yellow spheres indicate urea molecules. Arrows represent the inhomogeneous effect of pressure in contrast to that of urea.
Much debate regarding the contributions of cavities and hydration to the negative volume change upon unfolding, as well as compressibility effects, exists. Using NMR, the “squeezing” of the hydrogen bonds in parallel with the heterogeneous release of the water-excluded cavities by hydration was explored. High-precision data have allowed understanding these molecular events in detail. In contrast to the pushing effects of pressure, the authors show for the first time how urea acts along a protein, pulling its structure as it preferentially binds to the protein (Fig. 2). Taken together, these observations provide with mechanistic explanations of how urea controls protein unfolding. The results clearly establish the direct interaction mechanism as the dominant mechanism of urea-induced protein denaturation as shown by the peak intensities and systematic chemical shifts.
The ensemble of conformations populated at low urea concentrations is consistent with a DMG (dry molten globule) in which the protein surface is dominated by urea instead of water molecules. Tracing some parallels with high pressure, one can state that pressure affects protein structure due to compressibility effects leading to a shortening of protein–protein and protein–solvent H bonds: this phenomenon contrasts with the “H-bond shift” that occurs during urea binding (Fig. 2). Consequently, pressure ultimately pushes water molecules against protein atoms, heterogeneously affecting the structure, unlike urea, which homogenously pulls water molecules away from the solvation shell due to its binding properties. Water pull would drive the conformation into an unstable DMG state, thus explaining why small additional increases in urea concentration result in cooperative unfolding (Fig. 2). Similarly, pulling the structure would make it more sensitive to pressure, with a tendency to have larger volume changes. The separate and combined thermodynamic effects of urea and pressure have allowed providing a “push-and-pull” molecular interpretation for these effects, taking advantage of site-specific (NMR) and global (fluorescence and SAXS) methods. These interpretations provide insights that illuminate the cumulative effects of using subdenaturing concentrations of urea and pressure to measure protein folding and stability. In conclusion, the authors have presented experimental data introducing the “push-and- pull” hypothesis, which may explain differences between the molecular mechanisms involved during the physical and chemical unfolding of proteins. These observations open up possibilities for the study of protein folding and provide a new interpretation to explain the nature of the cooperative behavior of proteins during folding reactions.
Source: Guilherme A. P. de Oliveira and Jerson L. Silva, “A hypothesis to reconcile the physical and chemical unfolding of proteins”, PNAS 2015 112: E2775-E2784. DOI: 10.1073/pnas.1500352112
The studies of the size, structure and magnetic properties of the multifunctional brick-like $ \rm Ag@Fe_3O_4$ NPs obtained reveal them as possible candidates for advanced medical purposes.
The authors have demonstrated the effective biological activity of a noncomplex nanomaterial against susceptible and antibiotic-resistant bacteria without significant cytotoxicity.